Mitochondrial Eve
| Haplogroup Modern humans
|
|
| Possible time of origin | 152,000 - 234,000 BP[1] |
| Possible place of origin | East Africa |
| Ancestor | Neandertal-Human MRCA |
| Descendants | Mitochondrial macro-haplogroups L0, L1, and L5 |
|---|---|
| Defining mutations | None |
In the field of human genetics, Mitochondrial Eve refers to the most recent common matrilineal ancestor (MRCA) from whom all living humans are descended. Passed down from mother to offspring, all mitochondrial DNA (mtDNA) in every living person is directly descended from hers. Mitochondrial Eve is the female counterpart of Y-chromosomal Adam, the patrilineal most recent common ancestor, although they lived thousands of years apart.
Mitochondrial Eve is generally estimated to have lived around 200,000 years ago,[2] most likely in East Africa,[3] when Homo sapiens sapiens were developing as a species separate from other human species.
Mitochondrial Eve lived much earlier than the out of Africa migration that is thought to have occurred between 95,000 to 45,000 BP.[4] The dating for 'Eve' was a blow to the multiregional hypothesis, and a boost to the hypothesis that modern humans originated relatively recently in Africa and spread from there, replacing more "archaic" human populations such as Neanderthals. As a result, the latter hypothesis is now the dominant one.
|
Female and mitochondrial ancestry
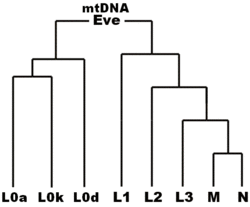
Without a DNA sample, it is not possible to reconstruct the complete genetic makeup (genome) of any individual who died very long ago. By analysing descendants' DNA, however, parts of ancestral genomes are estimated by scientists. Mitochondrial DNA (mtDNA) and Y chromosome are commonly used to trace ancestry in this manner. mtDNA is generally passed un-mixed from mothers to children of both sexes, along the maternal line, or matrilineally.[5][6] Matrilineal descent goes back to our mothers, to their mothers, until all female lineages converge.
Branches are identified by one or more unique markers which give a mitochondrial "DNA signature" or "haplotype" (e.g. the CRS is a haplotype). Each marker is a DNA base-pair that has resulted from an SNP mutation. Scientists sort mitochondrial DNA results into more or less related groups, with more or less recent common ancestors. This leads to the construction of a DNA family tree where the branches are in biological terms clades, and the common ancestors such as Mitochondrial Eve sit at branching points in this tree. Major branches are said to define a haplogroup (e.g. CRS belongs to haplogroup H), and large branches containing several haplogroups are called "macro-haplogroups".
The mitochondrial clade which Mitochondrial Eve defines is the species Homo sapiens sapiens itself, or at least the current population or "chronospecies" as it exists today. In principle, earlier Eves can also be defined going beyond the species, for example one who is ancestral to both modern humanity and Neanderthals, or, further back, an "Eve" ancestral to all members of genus Homo and chimpanzees in genus Pan. According to current nomenclature, Mitochondrial Eve's haplogroup was within mitochondrial haplogroup L because this macro-haplogroup contains all surviving human mitochondrial lineages today.
The variation of mitochondrial DNA between different people can be used to estimate the time back to a common ancestor, such as Mitochondrial Eve. This works because, along any particular line of descent, mitochondrial DNA accumulates mutations at the rate of approximately one every 3500 years.[7][8] A certain number of these new variants will survive into modern times and be identifiable as distinct lineages. At the same time some branches, including even very old ones, come to an end, when the last family in a distinct branch has no daughters.
Mitochondrial Eve is the most recent common matrilineal ancestor for all modern humans. Whenever one of the two most ancient branch lines dies out, the MRCA will move to a more recent female ancestor, always the most recent mother to have more than one daughter with living maternal line descendants alive today. The number of mutations that can be found distinguishing modern people is determined by two criteria: firstly and most obviously, the time back to her, but secondly and less obviously by the varying rates at which new branches have come into existence and old branches have become extinct. By looking at the number of mutations which have been accumulated in different branches of this family tree, and looking at which geographical regions have the widest range of least related branches, the region where Eve lived can be proposed.
The date when Mitochondrial Eve lived is estimated by determining the MRCA of a sample of mtDNA lineages. In 1980, Brown first proposed that modern humans possessed a mitochondrial common ancestor that may have lived as recently as 180kya. In 1987, Cann et al. suggested that mitochondrial Eve may have lived between 140-280kya.
Common fallacies
Not the only woman
One of the misconceptions of mitochondrial Eve is that since all women alive today descended in a direct unbroken female line from her that she was the only woman alive at the time.[9][10] However nuclear DNA studies indicate that the size of the ancient human population never dropped below some tens of thousands;[9] there were many other women around at Eve's time with descendants alive today, but somewhere in all their lines of descent there is at least one man (and men do not pass on their mothers' mitochondrial DNA to their children). By contrast, Eve's lines of descent to each person alive today includes at least one line of descent to each person which is purely matrilineal.
Not alive at the same time as "Adam"
Sometimes mitochondrial Eve is assumed to have lived at the same time as Y-chromosomal Adam, perhaps even meeting and mating with him. Like Eve, "Adam" probably lived in Africa; however, Eve lived much earlier than Adam – perhaps some 50,000 to 80,000 years earlier than Adam – due to the greater variability in male fecundity.[11]
Mitochondrial Eve is the most recent common matrilineal ancestor, not the most recent common ancestor (MRCA). Since the mtDNA is inherited maternally and recombination is either rare or absent, it is relatively easy to track the ancestry of the lineages back to a MRCA; however this MRCA is valid only when discussing mitochondrial DNA. An approximate sequence from youngest to oldest can list various important points in the ancestor of modern human populations:
- The Human MRCA. All humans alive today share a surprisingly recent common ancestor, perhaps even within the last 5000 years, even for people born on different continents.[12]
- The Identical ancestors point. Just a few thousand years before the most recent single ancestor shared by all living humans comes the time at which all humans who were alive either left no descendents or are common ancestors to all humans alive today. In other words, from this point back in time "each present-day human has exactly the same set of genealogical ancestors". This is far more recent than Mitochondrial Eve.[12]
- "Y-Chromosomal Adam", the most recent male-line ancestor of all living men, was much more recent than Mitochondrial Eve, but is also likely to have been long before the Identical ancestors point.
Implications of dating and placement of Eve
The first studies suggesting a recent common ancestor for humans within Africa came at time when a hypothesis for human evolution, known as the Multiregional Evolution hypothesis, was popular among some leading paleoanthropologists (e.g. Milford Wolpoff). The impetus for this hypothesis comes from the belief that humans first left Africa about 2 million years ago and spread globally; these humans were similar to modern humans in many ways. Other biases indicated the temporal difference between Homo erectus and modern Homo sapiens was too short to allow for another new species, and many authors perceived regional evolution from archaic forms into modern ones. Consequently, the finding of a recent maternal ancestor for all humans in Africa created an extended controversy. Over a decade the MREH theory retracted to theories of racial admixture, such as proposed by Eric Trinkhaus, to mostly recent Africa origin hypotheses.
The strict Out of Africa and Multiregional Evolution controversy revolved around where to best place the evolution of anatomically modern humans over evolutionary timescales.
Cann, Stoneking & Wilson (1987) placement of a relatively small population of humans in sub-saharan Africa, lent appreciable support for the recent Out of Africa hypothesis. The current concept places between 1,500 and 16,000 effectively interbreeding individuals (census 4,500 to 48,000 individuals) within Tanzania and proximal regions. Later, Tishkoff using data from many loci has extrapolated origins to the Angola-Namibia border region near the Atlantic Ocean (although this region has poor genetic definition), whereas Behar et al. 2008 places an ancestral population in Ethiopia. These opinions all point toward a sub-Saharan origin. More recent literature on languages and pygmy phenotype indicate that L0 and L1 were carried by click-speaking pygmies from SE Africa to Central and Western Africa, therefore explaining much of the genetic diversity in those regions. Consequently, more recent studies have tended to push the cradle of humanity more toward the South or East of Africa.
To some extent the studies have already revealed that the presence of archaic homo sapiens in Northwest Africa (Jebel Irhoud) were not likely part of the contiguous modern human population. In addition, the older remains at Skhul and Qafzeh are also unlikely part of the constrict human population, evidence currently indicates humans expanded in the region no earlier than 90,000 BP. Tishkoff argues that humans might have migrated to the Levant before 90 Ka, but this colony did not persist in SW Asia. Better defined is the genetic separation among Neanderthals, Flores hobbit, Java man, and Peking man. In 1999 Krings et al., eliminated problems in molecular clocking postulated by Nei, 1992 when it was found the mtDNA sequence for the same region was substantially different from the MRCA relative to any human sequence. Currently there are 6 fully sequenced Neanderthal mitogenomes, each falling within a genetic cluster less diverse than that for humans, and mitogenome analysis in humans has statistically markedly reduced the TMRCA range so that it no longer overlaps with Neandertal/human split times. Of all the non-African hominids European archaics most closely resembled humans, indicating a larger genetic divide with other hominids.
Since the Multiregional evolution hypothesis (MREH) revolved around a belief that regional modern human populations evolved in situ in various regions (Europe - Neandertals to Europeans, Asia - Homo Erectus to East Asians, Australia - Sumatran erectines to Indigeonous Australians), these results demonstrated that a pure MREH hypothesis could not explain one important genetic marker.
In popular science and culture
Popular science

Newsweek Magazine reported on Mitochondrial Eve based on the Cann et al. study in January 1988, under a heading of "Scientists Explore a Controversial Theory About Man's Origins". The edition sold a record number of copies.[13]
The Seven Daughters of Eve presents the theory of human mitochondrial genetics to a general audience.
In River Out of Eden, Richard Dawkins discusses human ancestry in the context of a river of genes and shows that Mitochondrial Eve is one of the many common ancestors we can trace back to via different gene pathways.
The Discovery Channel produced a documentary entitled The Real Eve (or Where We Came From in the United Kingdom), based on the book Out of Eden by Stephen Oppenheimer.
NOVA on PBS had an episode "Children of Eve" (about 1987).
Popular culture
- In his short story Mitochondrial Eve, Australian science fiction writer Greg Egan imagines the social consequences of the desire to identify, and identify with, one's haplogroup.
- The main villainess of the Parasite Eve series is named Mitochondria Eve.
- In the reimagined Battlestar Galactica, Hera Agathon (the Cylon-human hybrid) is the Mitochondrial Eve as revealed in the series finale.[14]
See also
|
|
|
Evolutionary tree of Human mitochondrial DNA (mtDNA) haplogroups |
||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | ||||||||||||||||||||||||||||||||
| L0 | L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||
| M | N | |||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | A | S | R | I | W | X | Y | |||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | |||||||||||||||||||||||||
| HV | JT | K | ||||||||||||||||||||||||||||||
| H | V | J | T | |||||||||||||||||||||||||||||
References
- ↑ Pedro Soares et al 2009, Correcting for Purifying Selection: An Improved Human Mitochondrial Molecular Clock. and its Supplemental Data. The American Journal of Human Genetics, Volume 84, Issue 6, 740-759, 04 June 2009
- ↑ University of Leeds - New ‘molecular clock’ aids dating of human migration history
- ↑ 'Your Genetic Journey' - The Genographic Project
- ↑ Endicott, P; Ho, SY; Metspalu, M; Stringer, C (September 2009), "Evaluating the mitochondrial timescale of human evolution", Trends Ecol. Evol. (Amst.) 24 (9): 515–21, doi:10.1016/j.tree.2009.04.006, PMID 19682765
- ↑ Giles, Richard E; H Blanc, H M Cann, and D C Wallace (1980), "Maternal inheritance of human mitochondrial DNA", PNAS 77 (11): 6715–6719, doi:10.1073/pnas.77.11.6715, PMID 6256757, PMC 350359, http://www.pnas.org/content/77/11/6715.abstract
- ↑ Birky, C. William (2008), "Uniparental inheritance of organelle genes", Current Biology 18 (16): R692–R695, doi:10.1016/j.cub.2008.06.049, PMID 18727899, http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6VRT-4T96DJ4-B&_user=10&_coverDate=08%2F26%2F2008&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1232071752&_rerunOrigin=scholar.google&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=b73147dffdd2ca6a010de407176efd84
- ↑ Soares, P; Ermini, L; Thomson, N; Mormina, M; Rito, T; Röhl, A; Salas, A; Oppenheimer, S et al. (June 2009), "Correcting for purifying selection: an improved human mitochondrial molecular clock", American Journal of Human Genetics 84 (6): 740–59, doi:10.1016/j.ajhg.2009.05.001, PMID 19500773
- ↑ There are sites in mtDNA (such as: 16129, 16223, 16311, 16362) that evolve more rapidly, have been noted to change within intragenerational timeframes - Excoffier & Yang (1999); however most studies avoid using these sites because of the higher possibly of reverse mutations causing information loss.
- ↑ 9.0 9.1 Takahata, N (January 1993), "Allelic genealogy and human evolution", Mol. Biol. Evol. 10 (1): 2–22, PMID 8450756, http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=8450756"Any hypothesis that assumes a small number of founding individuals throughout the late Pleistocene can be rejected."
- ↑ Dawkins, Richard (2004), The ancestor's tale: a pilgrimage to the dawn of evolution, Boston: Houghton Mifflin, ISBN 0-618-00583-8
- ↑ Mitochondrial Eve and Y-chromosomal Adam The Genetic Genealogist
- ↑ 12.0 12.1 Rohde, DL; Olson, S; Chang, JT (September 2004), "Modelling the recent common ancestry of all living humans", Nature 431 (7008): 562–6, doi:10.1038/nature02842, PMID 15457259
- ↑ Oppenheimer, Stephen (2004), The Real Eve: Modern Man's Journey Out of Africa, New York, NY: Carroll & Graf, ISBN 0-7867-1334-8
- ↑ The McGuffin Review: BATTLESTAR GALACTICA The Series Finale
Further reading
- Atkinson, QD; Gray, RD; Drummond, AJ (January 2009), "Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa", Proceedings. Biological Sciences / the Royal Society 276 (1655): 367–73, doi:10.1098/rspb.2008.0785, PMID 18826938, PMC 2674340, http://rspb.royalsocietypublishing.org/cgi/pmidlookup?view=long&pmid=18826938
- Ayala, F (1995), "The myth of Eve:molecular biolofy and human origin", Science 270 (5244): 1930–1936, doi:10.1126/science.270.5244.1930, PMID 8533083
- Balloux, F; Handley, LJ; Jombart, T; Liu, H; Manica, A (2009), "Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation.", Proc Biol Sci. 276 (1672): 3447–55, doi:10.1098/rspb.2009.0752, PMID 19586946
- Behar et al., D; Villems; Soodyall; Blue-smith; Pereira; Metspalu; Scozzari; Makkan et al. (May 2008), "The dawn of human matrilineal diversity", American Journal of Human Genetics 82 (5): 1130–40, doi:10.1016/j.ajhg.2008.04.002, PMID 18439549, PMC 2427203, http://linkinghub.elsevier.com/retrieve/pii/S0002-9297(08)00255-3
- Brown, WM (June 1980), "Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis", Proc. Natl. Acad. Sci. U.S.A. 77 (6): 3605–9, doi:10.1073/pnas.77.6.3605, PMID 6251473
- Cann, RL; Stoneking, M; Wilson, AC (1987), "Mitochondrial DNA and human evolution", Nature 325 (6099): 31–6, doi:10.1038/325031a0, PMID 3025745
- Cox, MP (August 2008), "Accuracy of molecular dating with the rho statistic: deviations from coalescent expectations under a range of demographic models", Hum. Biol. 80 (4): 335–57, doi:10.3378/1534-6617-80.4.335, PMID 19317593
- Dawkins, Richard (2004), The ancestor's tale: a pilgrimage to the dawn of evolution, Boston: Houghton Mifflin, ISBN 0-618-00583-8
- Dennett, Daniel Clement (1995), Darwin's dangerous idea: evolution and the meanings of life, New York: Simon & Schuster, ISBN 0-684-80290-2, http://books.google.com/books?id=FvRqtnpVotwC&printsec=frontcover#PPA98,M1
- Endicott, P; Ho, SY (April 2008), "A Bayesian evaluation of human mitochondrial substitution rates", Am. J. Hum. Genet. 82 (4): 895–902, doi:10.1016/j.ajhg.2008.01.019, PMID 18371929
- Endicott, P; Ho, SY; Metspalu, M; Stringer, C (September 2009), "Evaluating the mitochondrial timescale of human evolution", Trends Ecol. Evol. (Amst.) 24 (9): 515–21, doi:10.1016/j.tree.2009.04.006, PMID 19682765
- Excoffier, L; Yang, Z (October 1999), "Substitution rate variation among sites in mitochondrial hypervariable region I of humans and chimpanzees", Mol. Biol. Evol. 16 (10): 1357–68, PMID 10563016, http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=10563016
- Felsenstein, J (April 1992), "Estimating effective population size from samples of sequences: inefficiency of pairwise and segregating sites as compared to phylogenetic estimates", Genet. Res. 59 (2): 139–47, doi:10.1017/S0016672300030354, PMID 1628818
- Ferris, SD; Brown, WM; Davidson, WS; Wilson, AC (October 1981), "Extensive polymorphism in the mitochondrial DNA of apes", Proc. Natl. Acad. Sci. U.S.A. 78 (10): 6319–23, doi:10.1073/pnas.78.10.6319, PMID 6273863
- Gibbons, Anne (January 1998), "Calibrating the mitochondrial clock", Science 279 (5347): 28–9, doi:10.1126/science.279.5347.28, PMID 9441404, http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=9441404
- Gonder, MK; Mortensen, HM; Reed, FA; de Sousa, A; Tishkoff, SA (December 2007), "Whole-mtDNA genome sequence analysis of ancient African lineages", Mol. Biol. Evol 24 (3): 757–68, doi:10.1093/molbev/msl209, PMID 17194802
- Hazelwood, L; Steele, J (2004), "Spatial dynamics of human dispersals: Constraints on modelling and archaeological validation", Journal of Archaeological Science 31: 669–679, doi:10.1016/j.jas.2003.11.009, http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WH8-4BP3JJK-2&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&_docanchor=&view=c&_searchStrId=1143250278&_rerunOrigin=scholar.google&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=0cf2e2392a9cb119d6b6893d73727d66
- Ho, SY; Larson, G (February 2006), "Molecular clocks: when times are a-changin'", Trends Genet. 22 (2): 79–83, doi:10.1016/j.tig.2005.11.006, PMID 16356585, http://linkinghub.elsevier.com/retrieve/pii/S0168-9525(05)00335-5
- Ingman, M; Kaessmann, H; Pääbo, S; Gyllensten, U (December 2000), "Mitochondrial genome variation and the origin of modern humans", Nature 408 (6813): 708–13, doi:10.1038/35047064, PMID 11130070
- Kaessmann, H; Pääbo, S (January 2002), "The genetical history of humans and the great apes", J. Intern. Med. 251 (1): 1–18, doi:10.1046/j.1365-2796.2002.00907.x, PMID 11851860, http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=0954-6820&date=2002&volume=251&issue=1&spage=1
- Loogväli et al., Eva-Liis; Kivisild, Toomas; Margus, Tõnu; Villems, Richard; O'Rourke, Dennis (2009), "Explaining the Imperfection of the Molecular Clock of Hominid Mitochondria", PLoS ONE (PLoS ONE) 4 (12): e8260, doi:10.1371/journal.pone.0008260, PMID 20041137, PMC 2794369, http://www.plosone.org/article/info:doi/10.1371/journal.pone.0008260
- Kimura, Motoo; Ohta, Tomoko (2001), Theoretical Aspects of Population Genetics, Princeton University Press, pp. 232, ISBN 0691080984
- Loewe, L; Scherer, S (November 1997), "Mitochondrial Eve: The Plot Thickens", Trends in Ecology & Evolution 12 (11): 422–3, doi:10.1016/S0169-5347(97)01204-4
- Maca-Meyer, N; González, AM; Larruga, JM; Flores, C; Cabrera, VM (2001), "Major genomic mitochondrial lineages delineate early human expansions", BMC Genet. 2: 13, doi:10.1186/1471-2156-2-13, PMID 11553319, PMC 55343, http://www.biomedcentral.com/1471-2156/2/13
- Mishmar, D; Ruiz-Pesini, E; Golik, P; Macaulay, V; Clark, AG; Hosseini, S; Brandon, M; Easley, K et al. (January 2003), "Natural selection shaped regional mtDNA variation in humans", Proc. Natl. Acad. Sci. U.S.A. 100 (1): 171–6, doi:10.1073/pnas.0136972100, PMID 12509511
- Nei, M (November 1992), "Age of the common ancestor of human mitochondrial DNA", Mol. Biol. Evol. 9 (6): 1176–8, PMID 1435241, http://mbe.oxfordjournals.org/cgi/reprint/9/6/1176
- Nielsen, R; Beaumont, MA (March 2009), "Statistical inferences in phylogeography", Mol. Ecol. 18 (6): 1034–47, doi:10.1111/j.1365-294X.2008.04059.x, PMID 19207258
- Oppenheimer, Stephen (2004), The Real Eve: Modern Man's Journey Out of Africa, New York, NY: Carroll & Graf, ISBN 0-7867-1334-8
- Parsons, TJ; Muniec, DS; Sullivan, K; et al, N; Alliston-Greiner, R; Wilson, MR; Berry, DL; Holland, KA et al. (April 1997), "A high observed substitution rate in the human mitochondrial DNA control region", Nat. Genet. 15 (4): 363–8, doi:10.1038/ng0497-363, PMID 9090380
- Pritchard, JK; Seielstad, MT; Perez-Lezaun, A; Feldman, MW (1999), "Population growth of human Y chromosomes: a study of Y chromosome microsatellites..", Mol Biol Evol. 16 (12): 1791–98, PMID 10605120
- Reed, FA; Tishkoff, SA (2006), "Africa human diversity, origins and migrations", Current Opinions in Genetics & Development 16: 598
- Rohde, DL; Olson, S; Chang, JT (September 2004), "Modelling the recent common ancestry of all living humans", Nature 431 (7008): 562–6, doi:10.1038/nature02842, PMID 15457259
- Santos, C; Sierra, B; Alvarez, L; Ramos, A; Fernández, E; Nogués, R; Aluja, MP (2008), "Frequency and pattern of heteroplasmy in the control region of human mitochondrial DNA.", J Mol Evol. 67 (2): 191–200, doi:10.1007/s00239-008-9138-9, PMID 18618067
- Schaffner, SF (2004), "The X chromosome in population genetics", Nat Rev Genet 5 (1): 43–51, doi:10.1038/nrg1247, PMID 14708015, http://www.nature.com/nrg/journal/v5/n1/abs/nrg1247.html
- Soares, P; Ermini, L; Thomson, N; Mormina, M; Rito, T; Röhl, A; Salas, A; Oppenheimer, S et al. (June 2009), "Correcting for purifying selection: an improved human mitochondrial molecular clock", American Journal of Human Genetics 84 (6): 740–59, doi:10.1016/j.ajhg.2009.05.001, PMID 19500773
- Suissa, S; Wang, Z; Poole, J; Wittkopp, S; Feder, J; Shutt, TE; Wallace, DC; Shadel, GS et al. (2009), "Ancient mtDNA genetic variants modulate mtDNA transcription and replication.", PLoS Genet. 5 (5): e1000474, doi:10.1371/journal.pgen.1000474, PMID 19424428, PMC 2673036, http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.1000474
- Sykes, Bryan (2001), The Seven Daughters of Eve, New York: Norton, ISBN 0-393-02018-5
- Sykes, Brian D. (2003), Adam's curse: a future without men, London: Bantam, ISBN 0-593-05005-3
- Takahata, N (January 1993), "Allelic genealogy and human evolution", Mol. Biol. Evol. 10 (1): 2–22, PMID 8450756, http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=8450756
- Takahata, N; Lee, SH; Satta, Y (2001), "Testing multiregionality of modern human origins", Mol Biol Evol 18 (2): 172–183, PMID 11158376, http://mbe.oxfordjournals.org/cgi/content/full/18/2/172
- Tamura, K,; Nei, M (May 1993), "Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees", Mol. Biol. Evol. 10 (3): 512–26, PMID 8336541, http://mbe.oxfordjournals.org/cgi/reprint/10/3/512
- Tang, H; Siegmund, DO; Shen, P; Oefner, PJ; Feldman, MW (May 2002), "Frequentist estimation of coalescence times from nucleotide sequence data using a tree-based partition", Genetics 161 (1): 447–59, PMID 12019257, PMC 1462078, http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462078/pdf/12019257.pdf*Vigilant, L; Pennington, R; Harpending, H; Kocher, TD; Wilson, AC (December 1989), "Mitochondrial DNA sequences in single hairs from a southern African population", Proc. Natl. Acad. Sci. U.S.A. 86 (23): 9350–4, doi:10.1073/pnas.86.23.9350, PMID 2594772
- Vigilant, L; Stoneking, M; Harpending, H; Hawkes, K; Wilson, AC (September 1991), "African populations and the evolution of human mitochondrial DNA", Science 253 (5027): 1503–7, doi:10.1126/science.1840702, PMID 1840702, http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=1840702
- Watson E, Forster P, Richards M, Bandelt HJ (September 1997), "Mitochondrial footprints of human expansions in Africa", Am. J. Hum. Genet. 61 (3): 691–704, doi:10.1086/515503, PMID 9326335, PMC 1715955, http://linkinghub.elsevier.com/retrieve/pii/S0002-9297(07)64333-X
- Wilson, AC; Cann, RL; Carr, SM; George M, UB; Gyllensten, KM; Helm-Bychowski, RG; Higuchi, SR; Palumbi, EM et al. (1985), "Mitochondrial DNA and two perspectives on evolutionary genetics", Biol J Linn Soc Lond. 26 (4): 375–400, doi:10.1111/j.1095-8312.1985.tb02048.x, http://www3.interscience.wiley.com/journal/119851665/abstract
- Wilder, JA; Mobasher, Z; Hammer, MF (2004), "Genetic evidence for unequal effective population sizes of human females and males.", Mol Biol Evol. 21 (11): 2047–57, doi:10.1093/molbev/msh214, PMID 15317874
- White, TD; Asfaw, B; Beyene, Y; Haile-Selassie, Y; Lovejoy, CO; Suwa, G; WoldeGabriel, G (October 2009), "Ardipithecus ramidus and the paleobiology of early hominids", Science 326 (5949): 75–86, PMID 19810190
External links
- Krishna Kunchithapadam, "What, if anything, is a Mitochondrial Eve?" a simple explanation